- PDA –
-
potato dextrose agar
- PCA –
-
potato carrot agar
- ITS –
-
nuclear ribosomal internal transcribed spacer
- EF-1α –
-
translation elongation factor 1-alpha gene
- TUB2 –
-
beta-tubulin gene
- H3 –
-
histone 3 gene
- NaClO –
-
1% sodium hypochlorite solution
- ML –
-
Maximum likelihood tree
- UV –
-
ultraviolet
- CTAB –
-
cetyltrimethylammonium bromide
- NCBI –
-
the National Center for Biotechnology Information
Abbreviations
1. INTRODUCTION
There are 34 species in the genus Paeonia, which are grouped into two subgenera (Moutan and herbaceous peonies) and seven sections (Hong, 2021). Herbaceous peonies make up one of five sections that comprise the subgenus Paeonia (Prijić et al., 2023). Temperate Asia, southern Europe, and western North America are habitats for Paeonia peregrina, known as the Balkan or Kosovo peony, and Paeonia tenuifolia, known as the fern leaf or steppy peony ( The Plant List, 2023). In Serbia, their collection from nature is protected by law and requires permission from the Ministry of Environmental Protection of the Republic of Serbia.
Alternaria spp. can be found as saprophytic, endophytic, and pathogenic species in agricultural crops and products, soil, and organic matter (Neergaard, 1945; Pavón Moreno et al., 2012; Polizzotto et al., 2012). The species are located in semiarid and humid areas, and can affect leaves, stems, flowers, and fruits of several plant species (Deshpande, 2002). Also, Alternaria spp. are well-known as post-harvest pathogens that cause disease in over 100 host plants (Thomma, 2003). The species can survive on infected crop debris, seeds, and weeds in the form of mycelia or conidia, and can be transmitted by insects, wind, rain, irrigation, etc. Conidia are carried by air currents to the aerial parts of plants throughout the growing season (Battilani et al., 2009; Laemmlen, 2002). Alternaria spores are one of the most important airborne allergens (Thomma, 2003). The production of melanin, particularly in the spores, and the production of host-specific toxins in the case of pathogenic species are two distinguishing characteristics of Alternaria species (Thomma, 2003). The species of the genus, host plant, and combination of right temperature and humidity are important factors in the external environment for the development of infection. The spores penetrate the host’s tissue through stomata, cuticle, or wounds. The first symptoms of infection appear after 2–3 days on the leaves and stems of the host plant. Black necrotic lesions surrounded by chlorotic halos are characteristic of leaf spots. Leaf spot can lead to reduced leafy crops. Additionally, it may cause the host to abscise leaves, thus lowering photosynthetic potential and crop yield indirectly (Armitage, 2013; Battilani et al., 2009).
Despite the increase in peony production and consumption, available information on peony diseases and their treatment is limited. Alternaria spp. has been found on herbaceous and tree peonies in Canada (Coulson, 1923), the United States (Anonymous, 1960), China (Zhang, 2003), Korea (Cho and Shin, 2004), Bulgaria (Bobev, 2009), and the Netherlands (Andersen et al., 2009). The Texas Plant Disease Handbook (Anonymous, n.d.) describes the leaf spots as irregularly shaped, purplish-brown to reddish-brown. Later, the leaves may turn yellow and fall off (Texas Plant Disease Handbook). Coulson (1923) described Alternaria spp. as causing yellow leaf spots, irregularly shaped with a clearly defined, dark margin. Leaf spots caused by Alternaria suffruticosae, A. suffruticosicola, and A. tenuissima were circular or irregularly shaped, brown to black (Zhang et al., 2008). Cheng et al. (2018) recorded diversity of fungi of Aspergillus, Alternaria, and Penicillium on the leaf, stem, and root of Paeonia lactiflora Pallas. Alternaria sp., especially A. alternata, showed the maximum frequency of colonization and isolation (10.95% and 31.68%), similar to the results of previous reports (Jennings et al., 2002; Lin et al., 2007; Xiao et al., 2013). In addition, A. tenuissima was identified as the causal agent of red leaf spot disease on P. lactiflora in China (Sun and Huang, 2017). The first symptoms on the leaves appeared as small, circular, reddish to dark brown, necrotic spots that gradually enlarged and became irregular. The spots grew together, and the leaves withered, dried up, and defoliated. This disease destroys the aesthetic value of the Chinese peony and potentially reduces the yield and quality of its medicinal properties, affecting the economic importance of this plant.
The identification of Alternaria species is difficult and at present, molecular and phylogenetic analyses are essential for recognizing species (Lawrence et al., 2015; Woudenberg et al., 2015). Different loci have been used for molecular identification and characterization, among which are the nuclear ribosomal internal transcribed spacer (ITS) region (Lawrence et al., 2015; Matić et al., 2019; Woudenberg et al., 2015), partial translation elongation factor 1-alpha (EF-1α) (Lawrence et al., 2015; Siciliano et al., 2018; Woudenberg et al., 2015), beta-tubulin (TUB2) (Lawrence et al., 2015; Matić et al., 2019; Peever et al., 2004; Siciliano et al., 2018), and histone 3 gene (H3) (Garibaldi et al., 2018a; Matić et al., 2019; Wang et al., 2019; Zhang et al., 2020; Zhao et al., 2016).
In Serbia, A. alternata has been reported on P. peregrina originating from the location of Bogovo Guvno (Mikić et al., 2023), while there isn‘t information about this pathogen on P. tenuifolia (Farr and Rossman, 2023).
2. MATERIALS AND METHODS
2.1. Plant collection and fungal isolation
In July 2021, leaf spot were observed on P. peregrina and P. tenuifolia in the natural habitats in Pirot (43◦ 07’ N; 22◦ 26’ E 666) and Deliblato Sands (44◦ 57’ N; 21◦ 03’ E 167) areas, Serbia. The spots appeared on leaves and sporadically on stems of P. peregrina, while identical spots, white mycelia and rot appeared on stems and lower branches of P. tenuifolia. The percentage of infected plants at a given locality were calculated by counting the numbers of healthy and symptomatic plants separately. In total, 197 and 100 individual plants of the single populations of P. peregrina and P. tenuifolia were found on about 0.5 ha. Sixty-three plants of P. peregrina and 50 plants of P. tenuifolia were symptomatic.
Symptomatic plants were transplanted into pots and transferred to the laboratory. Isolation of the pathogen was done from symptomatic leaves and stems. Leaf fragments (5x5 mm) of five symptomatic plants and stem fragments (5x5 mm) of ten symptomatic plants were cut from the margins of the lesions and the healthy tissue. Leaf and stems fragments were washed, surface disinfected with 75% ethanol for 1 min, 1% sodium hypochlorite solution (NaClO) for 2 min, rinsed three times in distilled water, dried on sterilized filter paper, and placed on potato dextrose agar (PDA) in Petri dishes (90 mm) (Sun et al., 2022). After seven days of incubation at 25 ◦C, developed colony fragments were transferred to PDA to obtain pure cultures.
2.2. Pathogenicity test
The pathogenicity was tested by artificial inoculation leaves of plant origin. Two wounded and two non-wounded detached leaves of healthy P. peregrina obtained from the natural habitat were used. The leaves were placed onto the filter paper moistened in Petri dishes, wounded with a sterile needle, and mycelial fragments (5 mm) of 7-day-old culture on PDA were placed on the adaxial surface of the leaves. Control leaves were treated the same way, but sterile PDA fragments were used (Fang et al., 2023). Petri dishes with inoculated leaves and control dishes were incubated at 21±2 ◦C under natural daylight conditions. The test was repeated once. Re-isolation of the pathogen was performed as described above.
2.3. DNA extraction, PCR amplification and amplicon sequencing
DNA extraction was performed by the cetyltrimethylammonium bromide (CTAB) protocol proposed by Day and Shattock (1997) from 7-day-old cultures grown on PDA. For molecular identification and characterization, the nuclear ribosomal internal transcribed spacer (ITS) region, partial translation elongation factor 1-alpha (EF-1α), beta-tubulin (TUB2), and histone 3 gene (H3) were amplified using primer pairs ITS1/ITS4 (White et al., 1990), EF1-728F/EF1-986R (Carbone and Kohn, 1999), T1/Bt2b (Glass and Donaldson, 1995; O’Donnell and Cigelnik, 1997), and H3-1a/H3-1b (Glass and Donaldson, 1995), respectively. Each 25 μL PCR mix contained 1 μL of template DNA, 1xPCR Master Mix (Thermo Scientific, Vilnius, Lithuania) and 0.4 μM of each primer. Conditions for amplification of ITS were: initial denaturation at 95 ◦C for 2 min, followed by 35 cycles of denaturation at 95 ◦C for 30 s, annealing at 55 ◦C for 30 s and elongation at 72 ◦C for 1 min. Conditions for EF-1α and TUB2 amplifications were: initial denaturation at 95 ◦C for 8 min, followed by 35 cycles of denaturation at 95 ◦C for 15 s, annealing at 55 ◦C for 20 s and elongation at 72 ◦C for 1 min. Conditions for H3 amplification were: initial denaturation at 94 ◦C for 2 min, denaturation at 94 ◦C for 60 s, annealing at 58 ◦C for 1 min and elongation 72 ◦C for 1 min; repeat protocol for 32 cycles. Final elongation occurred for 10 min at 72 ◦C. PCR products (5 μL) were observed in 1.2% agarose gel, stained in ethidium bromide, and visualized with UV transilluminator. Amplified products were purified and commercially sequenced (Macrogen Inc., Seoul, South Korea) using the same reverse primers as for amplification. The obtained sequences were assembled using Pregap4 from the Staden programme package (Staden et al., 1998). For species identification, the obtained sequences were compared with those publicly available in NCBI’s GenBank database using MegaBLAST (http://www.ncbi.nlm.nih.gov/). Sequences for each locus were aligned using Clustal X (Thompson, 1997), under MEGA version 7 (Kumar et al., 2016). The sequences of isolate were deposited in the National Center for Biotechnology Information (NCBI) GenBank.
2.4. Phylogenetic analyses
Evolutionary history was inferred based on combined analyses of the two (ITS and EF-1α) loci of two representative isolates obtained in this study, reference isolates of Alternaria spp. and Alternaria alternantherae CBS 124392 as an outgroup,using Maximum likelihood (ML) method (MEGA X) (Kumar et al., 2018). Sequences used for phylogeny are presented in Table 1. For the ML, the best nucleotide substitution model was determined using the “find best model” option in MEGA X. For tree inference, the nearest-neighbor-interchange heuristic method was employed, with the initial trees being automatically generated by Neighbor-Join and BioN Jalgorithms. To estimate the statistical significance of the inferred clades, each tree was bootstrapped 1,000 times.
| Isolate | Host/Location | GenBank accession numbers | Reference | |
|---|---|---|---|---|
| ITS | EF-1α | |||
| Alternaria alternata | ||||
| PpPili | Paeonia peregrina/Pirot | OP562152 | OP643688 | This study |
| SBDPst | Paeonia tenuifolia/Deliblatska peščara | nd | nd | This study |
| CBS 916.96T | Arachis hypogaea, India | AF347031 | KC584634 | (Woudenberg et al., 2015) |
| CBS 115188 | Citrus clementina, South Africa | KP124349 | KP125125 | (Woudenberg et al., 2015) |
| CBS 113013 | Malus domestica, South Africa | KP124341 | KP125117 | (Woudenberg et al., 2015) |
| CBS 117143 | Capsicum annuum, Italy | KP124355 | KP125131 | (Woudenberg et al., 2015) |
| CBS 119115 | Prunus sp., Greece | KP124360 | KP125136 | (Woudenberg et al., 2015) |
| CBS 918.96 (ex A. tenuissima R) | Dianthus chinensis, UK | AF347032 | KC584693 | (Woudenberg et al., 2015) |
| CBS 115152 | Psychotria serpens, China | KP124348 | KP125124 | (Woudenberg et al., 2015) |
| REIS 68 | Ceratostigma willmottianum, Italy | MN565914 | MN627329 | (Matić et al., 2020) |
| IT44 | Echinacea purpurea, Italy | MG182428 | MN627319 | (Matić et al., 2020) |
| IT61 | Mentha × piperita, Italy | MF997592 | MN627337 | (Matić et al., 2020) |
| Alternaria alstromeriae | ||||
| CBS 118809T | Alstroemeria sp., Australia | KP124297 | KP125072 | (Woudenberg et al., 2015) |
| CBS 118808R | Alstroemeria sp., USA | KP124296 | KP125071 | (Woudenberg et al., 2015) |
| Alternaria alternantherae | ||||
| CBS 124392 | Solanum melongena, China | KC584179 | KC584633 | (Woudenberg et al., 2015) |
| Alternaria arborescens | ||||
| CBS 102605T | Solanum lycopersicum, USA | AF347033 | KC584636 | (Woudenberg et al., 2015) |
| CBS 109730 | Solanum lycopersicum, USA | KP124399 | KP125177 | (Woudenberg et al., 2015) |
| Alternaria gaisen | ||||
| CBS 632.93R | Pyrus pyrifolia, Japan | KC584197 | KC584658 | (Woudenberg et al., 2015) |
| CBS 118488R | Pyrus pyrifolia, Japan | KP124427 | KP125206 | (Woudenberg et al., 2015) |
| Alternaria longipes | ||||
| CBS 540.94R | Nicotiana tabacum, USA | AY278835 | KC584667 | (Woudenberg et al., 2015) |
| CBS 121332R | Nicotiana tabacum, USA | KP124443 | KP125222 | (Woudenberg et al., 2015) |
Legend: T - ex-type isolate; R - representative isolate; nd - not deposited.
2.5. Morphological characterization
Macromorphological characteristics of two representative isolates (colony color, texture, and growth rate) were observed on PDA and PCA 40 cm below cool white fluorescent illumination at 23 ◦C for seven days (Blagojević, 2020; Simmons, 2007). The colony diameter was measured every day in two diagonal directions. Mean values were obtained for every day from three replicates with three plates each. The growth rate was calculated and expressed as the diameter of the colony after seven days. Micromorphological characteristics of selected isolates (length, width, shape, size, color of conidia, conidiophores, dimensions of the conical beak, and sporulation pattern) were observed on seven-day-old cultures grown on PCA at 22 ◦C under cool white fluorescent illumination and an 8/16 h light/dark regime (Simmons, 2007). Microscopic preparations were prepared in lactic acid and viewed under a microscope at 10x and 20x magnifications (Olympus CX43, Olympus, Hamburg, Germany). Photographs of conidiophore and conidia and measurements were obtained using the camera Axiocam ERc 5s, Zeiss, and software ZEN 2 (blue edition), Jena, Germany.
3. RESULTS AND DISCUSSION
3.1. Symptoms, isolates and pathogenicity
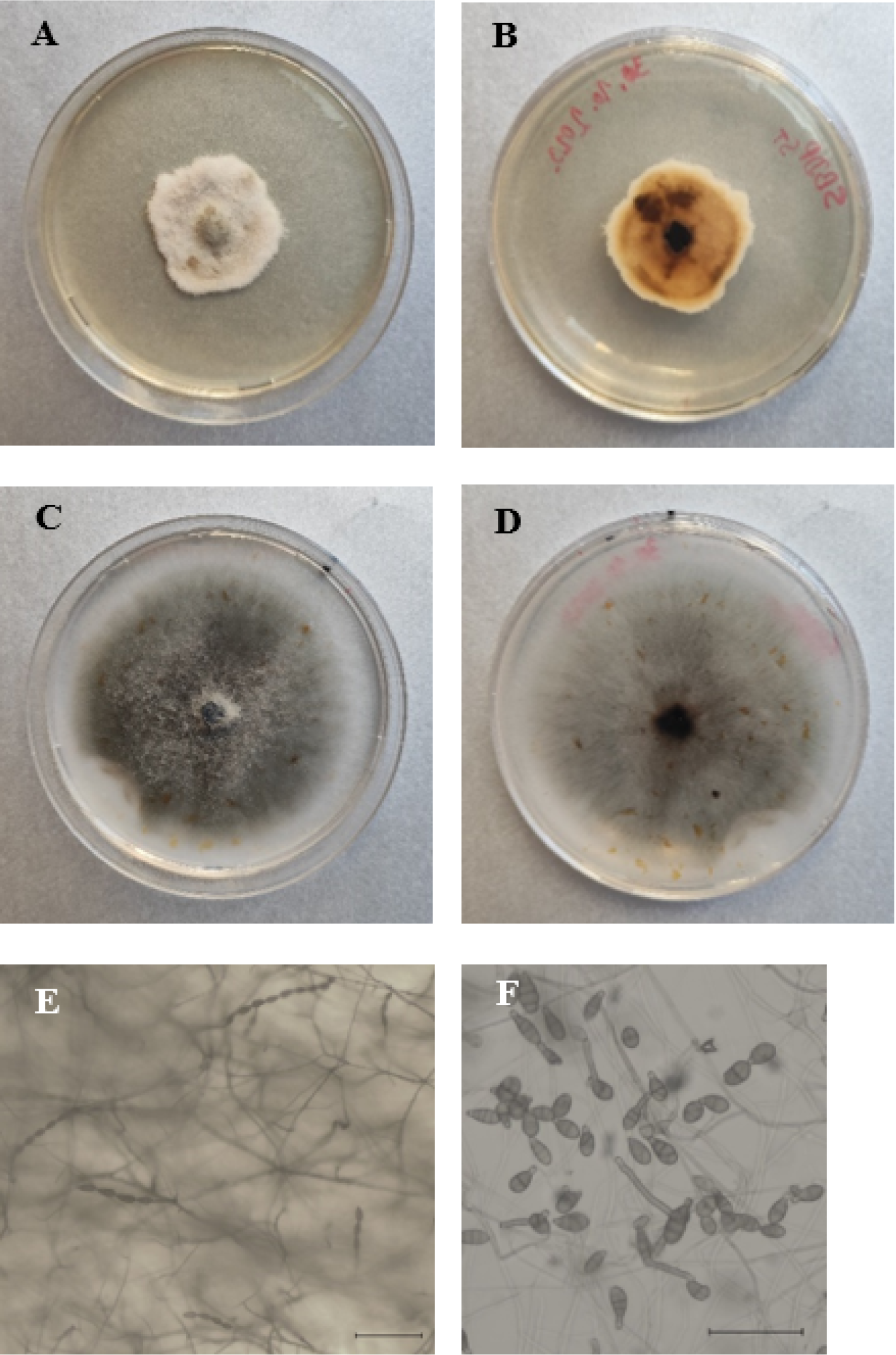
Circular or irregular, light to dark brown spots with defined dark margins appeared on the leaves and sporadically stems of P. peregrina during the stage of follicle formation, whereas on P. tenuifolia identical spots first appeared from the base at the bud stage and finally white mycelia and rot appeared at the base of the stems and lower branches at the seed maturity stage (Figure 1). The spread of spots on leaves causing the plants to wilt and die. Disease incidences in the areas of Pirot and Deliblato Sands were 32% and 25-35%, respectively.
Following a seven-day incubation, five isolates of P. peregrina and five isolates of P. tenuifolia were obtained from symptomatic plants. In the pathogenicity test, on wound-inoculated leaves, leaf necrosis and mycelia appeared on the adaxial surface of the leaves, as well as necrosis along the leaf nerve, while non-wounded leaves and control leaves remained symptomless (Figure 2). The pathogen was reisolated from inoculated leaves and showed identical morphological characteristics as the original isolate, thus completing Koch’s postulates. From each host, a representative isolate with proven pathogenicity was selected for further molecular and morphological characterization (isolate PpPili from P. peregrina and isolate SBDPst from P. tenuifolia).
3.2. Molecular identification
Following the amplification of ITS, EF-1α, TUB2 and H3, PCR products of expected size (600, 300, 600 and 400 bp, respectively) were obtained for the two selected isolates. Comparison of the obtained sequences showed that isolates PpPili and SBDPst were 100% similar (identical) in ITS and EF-1α, but differed in 1 nt in TUB2 and 1 nt in H3 gene regions. BLAST analysis showed that both isolates were 100% similar with A. alternata ex-type CBS 916.96, established by Woudenberg et al. (2013) and Woudenberg et al. (2015), in ITS (AF347031) and EF-1α (KC584634). Also, sequences of both regions of obtained isolates were identical with several corresponding sequences of A. alternata isolates (e.g. CBS 115188, CBS 113013, CBS 117143, CBS 119115, CBS 918.96, IT44, IT61) (Matić et al., 2020; Woudenberg et al., 2015). In TUB2, isolate PpPili was 100% similar with A. alternata ex-type CBS 916.96 (=EGS 34.016; MF070244) (Siciliano et al., 2018) and other published A. alternata sequences of TUB2 region (Garibaldi et al., 2019; 2020), while isolate SBDPst was 99% similar with A. alternata ex-type CBS 916.96 and 100% similar with CBS 918.96 and CBS 115152 (Siciliano et al., 2018). In H3, isolate PpPili was 100% similar with A. alternata isolate IT44 (MG182429) (Garibaldi et al., 2018a), while isolate SBDPst was 100% similar with A. alternata isolate IT61 (MF997593) (Garibaldi et al., 2018b). Based on ITS, EF-1α, TUB2 and H3, obtained isolates were identified as A. alternata which is in accordance with Woudenberg et al. (2015), Garibaldi et al. (2019; 2020; 2018a), Siciliano et al. (2018), and Matić et al. (2020).
3.3. Phylogeny
The phylogenetic analysis, based on Maximum likelihood method and Kimura 2-parameter model, of concatenated ITS and EF-1α sequences of Alternaria spp. grouped two representative isolates, PpPili and SBDPst, with reference isolates of A. alternata (Figure 3), confirming the identification.
3.4. Macromorphology and micromorphology
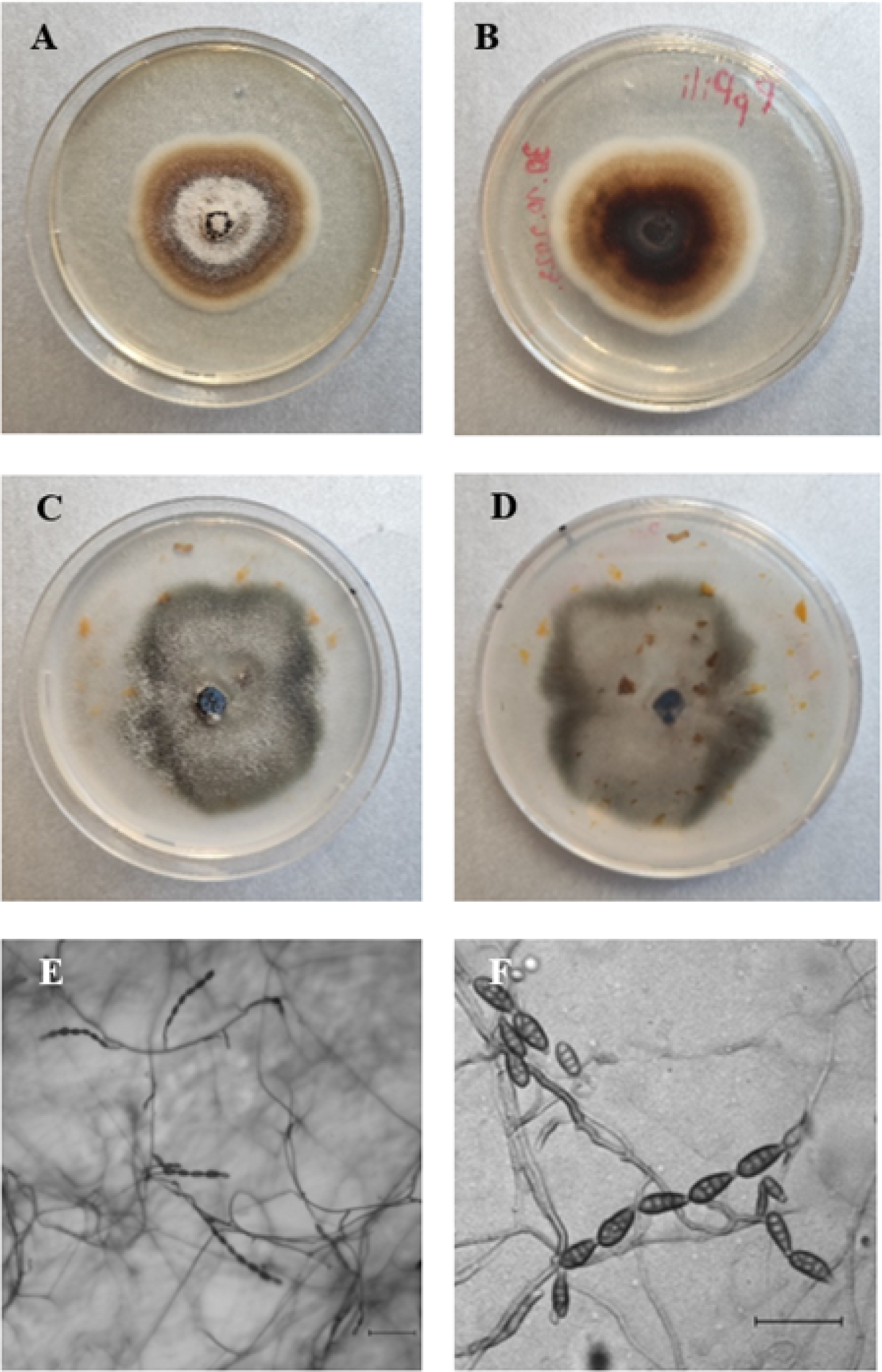
After seven days, isolate PpPili formed circular, cottony, light brown to brown colonies, with a white border on PDA (diameter 60.3 mm), and circular, grayish-brown colonies on PCA (diameter 83.7 mm). Conidia were brown, ovoid, or ellipsoid with three to four transverse and one to three longitudinal septa (16.01-27.74 x 5.50-14.41 μm; average 20.83 x 9.41 μm, n = 50). Conidia formed in chains of 2-7, mostly 5-6. Dimensions of the conical beak were 0.70-5.27 x 1.53-4.57 μm; average 2.55 x 2.68 μm, n = 50. Conidiophores were simple, dark brown, and unbranched (27.86-187.31 x 1.91-7.73 μm; average 59.88 x 4.39 μm, n = 50) (Figure 4).
After seven days, isolate SBDPst formed circular, cottony, white, or light brown colonies on PDA (diameter 45.3 mm). On PCA, colonies were circular, grayish-brown (diameter 85.5 mm), and brown, ovoid or ellipsoid conidia formed in chains of 3-10, mostly 3-8. Conidia had zero to four transverse and zero to three longitudinal septa, and were 13.56-24.27 x 5.7-14.3 μm in size (average 18.93 x 9.67 μm, n = 50), with a conical beak 1.00-5.52 in long (average 2.82) x 0.65-3.34 in wide (average 1.73) μm (n = 50). Conidiophores were simple, dark brown, and unbranched, 12.64-53.15 x 1.29-5.97 μm in size (average 25.98-2.36, n = 50) (Figure 5).
Observed morphological characteristics corresponded to the genus Alternaria (Simmons, 2007). According to the sporulation pattern the isolates could be identified as an A. tenuissima morphospecies, but with regards to a comprehensive revision by Woudenberg et al. (2015) and recently, Dettman and Eggertson (2021) which show lack of phylogenetic divergence in support of morphological differences and the fact that morphospecies are synonymized with A. alternata, the isolates were identified as A. alternata.
In this study, Alternaria leaf spot was described on two herbaceous peony species P. peregrina and P. tenuifolia in Serbia. Isolated fungi whose pathogenicity was proven, were identified as A. alternata. In Serbia, there has been a report of Alternaria leaf spot affecting wild herbaceous peony (Mikić et al., 2023), but other diseases have not been identified in these plant species. In the United States, Garfinkel and Chastagner (2019) described Alternaria leaf spot and other diseases on the herbaceous peony species P. lactiflora Pallas.. Also, Park et al. (1997) examined the effect of different pathogenic fungi (Alternaria sp., Erysiphe aquilegiae, and Cronartium flaccidum) on the growth and root yield of P. lactiflora; brown, dark-brown to black spots were observed on leaves, and finally the withering of the above-ground part of the plant. The symptoms of A. alternata on tree peonies described by Shi et al. (2015) were similar with our findings of A. alternata on herbaceous P. peregrina. Zhang et al. (2008) reported A. suffruticosae, A. suffruticosicola, and A. tenuissima on woody peony – Paeonia suffruticosa cultivar Andrews.
4. CONCLUSION
Alternaria spp. are present in humid and semi-arid regions, and can infect a wide range of plant crops through leaves, stems, flowers, and fruits. The fungus was isolated from infected plants collected at the localities Pirot and Deliblato Sands, and identified as A. alternata based on molecular characteristics. The ML phylogenetic tree of concatenated ITS and EF-1α sequences, and ITS, EF-1α, and TUB2 sequences of Alternaria spp., showed that two isolates obtained in this study clustered among A. alternata. After the pathogenicity test, the pathogen was reisolated from inoculated leaves and showed identical morphological characteristics as the original isolate, thus completing Koch’s postulates. Based on the observed morphological characteristics and sporulation pattern the isolates can be identified as A. tenuissima morphospecies, but this morphospecies in recent taxonomy has been synonymized with A. alternata. In Serbia, to the best of our knowledge, this is the first report of A. alternata infecting herbaceous wild peony species. Further investigations of A. alternata should elucidate the importance of the disease it causes on peonies and propose possible control measures to preserve this important, rare, and protected plant species in its natural habitats in Serbia.